Discover the ethical implications and breeding guidelines for genetically modified and genome-edited dairy cattle. How will these advancements shape the future of dairy farming?
Summary: Genetic modification and genome editing have revolutionized agricultural practices, offering unprecedented possibilities for enhancing dairy cattle traits. These technologies bring not only the promise of increased productivity and disease resistance but also complex ethical questions that must be addressed. Genetically modified (GM) and genome-edited dairy cattle are revolutionizing agriculture by introducing healthier, more productive, and ecologically friendly animals. The CRISPR-Cas9 technology is the most widely used genetic engineering approach, requiring continuous monitoring of the herd’s genetic health before and after genome editing. Breeding guidelines for genome-edited dairy calves must adhere to best practices, such as maintaining a varied gene pool to minimize inbreeding and disease susceptibility. However, negative genetic associations with milk production features hinder the development of udder health traits. Genetically engineered calves that produce recombinant human lactoferrin, lysozyme, or HBD-3 in milk have been developed, with studies showing that transgenic cows have fewer symptoms and cleared germs quicker than nontransgenic control cows. Ethical concerns surrounding GM and genome editing in dairy cattle include tampering with nature’s course, potential welfare consequences for animals, and potential effects on biodiversity.
- Genetic modification and genome editing are transforming dairy farming by enhancing traits like productivity and disease resistance.
- CRISPR-Cas9 is the prevalent technology used in genetic engineering, necessitating diligent herd genetic health monitoring.
- Best breeding practices for genome-edited dairy calves include maintaining genetic diversity to prevent inbreeding and reduce disease vulnerability.
- Negative genetic correlations with milk production traits can impede improving udder health.
- Transgenic cows can produce beneficial proteins such as recombinant human lactoferrin, lysozyme, or HBD-3, which have shown health advantages in research studies.
- Ethical considerations involve concerns about manipulating natural processes, animal welfare implications, and impacts on biodiversity.
The introduction of genetically modified (GM) and genome-edited dairy cattle is set to transform agriculture in ways we never imagined. Scientists strive to create a future where dairy cattle are healthier, more productive, and ecologically friendly through genetic modification. This shift from traditional breeding to cutting-edge genetic technology prompts us to ponder the complexities and implications for farmers, consumers, and animals. As we delve into this topic, we must grapple with the intriguing issues of science and technology and the intricate ethical perspectives that envelop it. This post encourages readers to engage with these issues and approach them with a sense of responsibility and thoughtfulness. Let’s embark on this thought-provoking journey together.
Understanding Genetic Modification and Genome Editing in Dairy Cattle
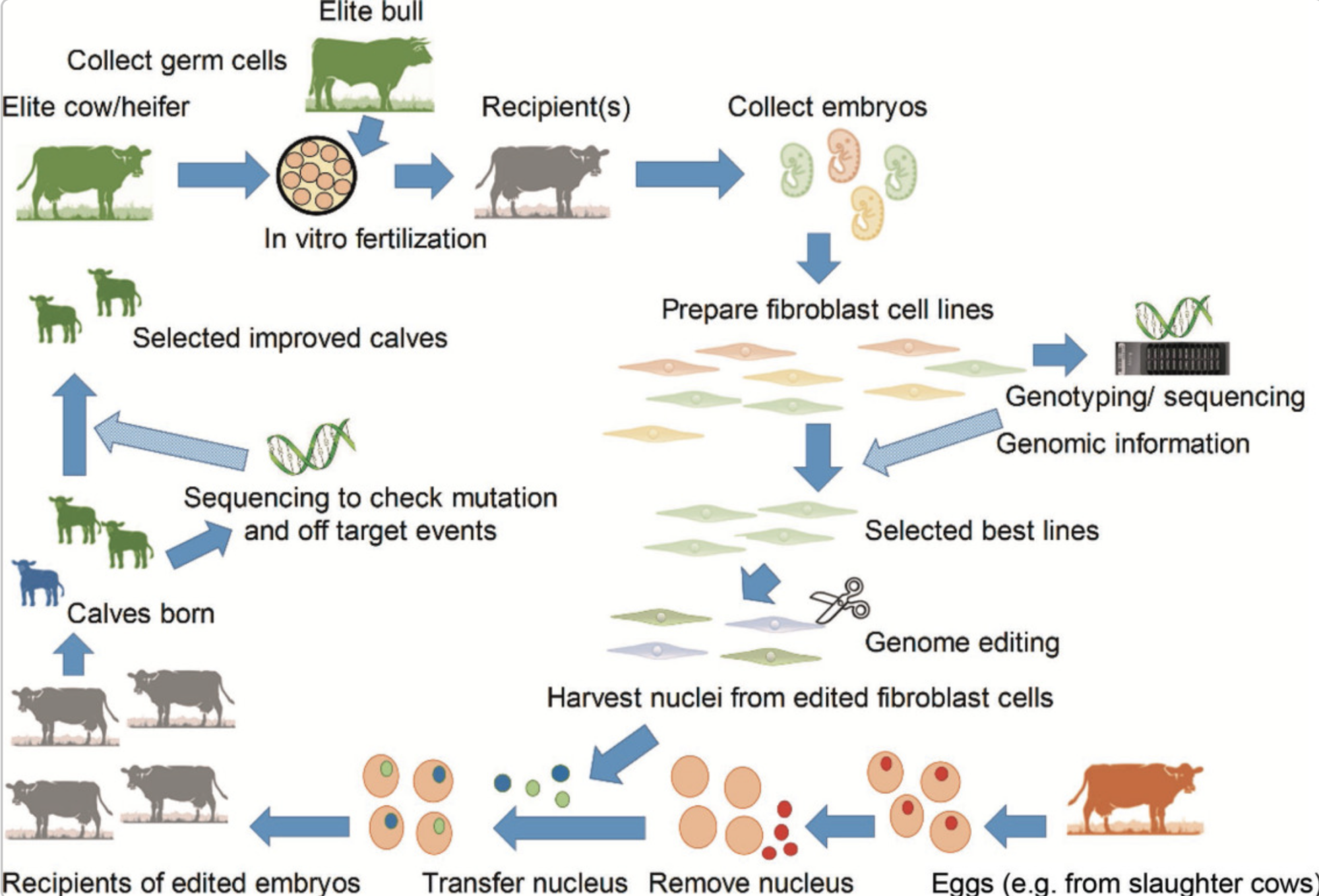
Consider the enormous possibilities for genetic manipulation and genome editing in dairy cattle. Consider animals that can generate lactose-free milk while being nutrient-dense and disease-resistant. This is not fiction; genetic engineering is a fast-emerging topic in animal production. Two basic genetic engineering approaches are in use today: transgenic and cisgenic. Transgenic refers to importing genes from one species into another, such as putting a bacterial gene into a cow’s genome. Conversely, Cisgenic entails changing a cow’s genes using genes from the same or nearly related species, similar to an enhanced form of conventional breeding techniques.
Today’s most extensively used approach for genome editing is the revolutionary ‘CRISPR-Cas9 technology.’ This groundbreaking tool allows scientists to modify gene sequences in a dairy cow’s DNA as easily as editing a page using a word processor. By using a scissor-like enzyme called Cas9, scientists can cut DNA strands at exact locations where alterations are required. The cell’s repair mechanism then takes charge, inserting or replacing genetic material to change the genome. This technology has the potential to revolutionize dairy cattle breeding.
To put this into perspective, consider a dairy cow with a genetic feature that makes it susceptible to a specific illness. Scientists may use genome editing to replace the disease-prone genetic sequence with one that increases resistance. The result is a healthier, more resilient, more productive dairy cow. This fantastic technology marks a considerable step in improving cattle welfare and agricultural efficiency.
Breeding Guidelines for Genome Edited Dairy Cattle: Best Practices
Breeding standards for genome-edited dairy calves must adhere to best practices to guarantee ethical and efficient operations. Continuous monitoring of the herd’s genetic health by tracking changes before and after genome editing and maintaining a varied gene pool to minimize inbreeding and disease susceptibility are critical steps toward ensuring the long-term viability of genome-edited cattle.
The following are some use cases for Genome Editing in Dairy Cattle:
- Case 1: Genome Editing to Eliminate Dehorning
Genetic dehorning of cattle is one possible use of genome editing in large-scale farming. Polledness, or the lack of horns, is an autosomal dominant feature involving two separate mutations in cow breeds. Dehorning is a routine practice to avoid accidents. Still, it is expensive and time-consuming, with over 80% of European dairy cattle dehorned without pain relief medication. However, this technique may produce quantifiable pain-related responses in cattle, prompting animal welfare issues. Although many cow herds include genetically polled breeding males, the number of polled AI breeding bulls in the Holstein breed still needs to be higher. Genome editing has been offered as a shortcut for producing high-quality polled bulls while minimizing genetic gain losses and using closely related polled individuals. Genome editing would generate a significant percentage of homozygous animals with the beneficial allele, raising allele frequency in the population. Selective matings between horned, homozygous, and heterozygous polled breeding bulls and cows might increase the number of polled calves produced. The first reported examples of genome-edited polled calves were created via SCNT, allowing the selection of embryos with specified changes before embryo transfer into the recipient cow. To effectively use genome editing to enhance the frequency of polled cattle, the sires and dams of edited embryos must have high genetic quality and be as unrelated as feasible. Large-scale breeding operations would utilize a mix of naturally polled, genome-edited polled, and dehorned breeding animals. - Case 2: Insertion of Human Genes to Increase Udder Health in Dairy Cattle
Udder health is critical for dairy output and animal welfare, and mastitis is a significant cause for culling in contemporary dairy herds. Genetic engineering (GM) has been utilized to enhance udder health by using indicator features such as milk SCC, which are more straightforward to evaluate continually. However, negative genetic associations with milk production features impede the development of udder health traits. There are many possible genes for mastitis resistance or susceptibility, including polymorphisms in genes that encode bovine lactoferrin and lysozyme. Lactoferrin concentration in bovine milk has a heritability of 0.22, indicating that genetic selection for higher lactoferrin levels is conceivable. However, the complexities of mastitis resistance persist, and appropriate bovine mastitis management is still missing. Genetically engineered calves that produce recombinant human lactoferrin, lysozyme, or HBD-3 in milk have previously been developed. According to studies, transgenic cows that generated recombinant human lactoferrin in their milk got infected with Staphylococcus chromogenes but had fewer symptoms and cleared germs quicker than nontransgenic control cows. GM cows expressing HBD3 or human lysozyme in milk seemed more resistant to bacterial udder infections than nontransgenic controls. In addition to improving udder health in dairy cows, generating bioactive recombinant human lactoferrin, lysozyme, and other agents in milk may benefit the gastrointestinal health of humans.
Ethical Dilemmas Surrounding Genetically Modified Dairy Cattle
While the advantages of utilizing genetic modification and genome editing in dairy cows are apparent, they are not without ethical implications. The idea of tampering with nature’s course typically raises eyebrows, and opponents are concerned about the possible welfare consequences for the animals themselves. Furthermore, there is worry about the potential effect on biodiversity, particularly if genetically modified creatures interbreed with non-modified ones. These issues are genuine and must be addressed to ensure the continuing development of this technology. However, these novel approaches have the potential to feed a rising global population in a sustainable, healthy, and efficient manner, which may eventually outweigh the possible concerns.
Ethical advisory committees inside breeding organizations may avoid gradual modifications that might result in a “slippery slope” effect. Instead of imposing extra restrictions, these committees should encourage internal conversations and decision-making. Implementing such organizations should not be treated lightly; they must address critical ethical concerns unique to each company to stay successful and productive. Successful ethical committees include the Dutch-Flemish cattle improvement cooperation CRV and worldwide pig breeding enterprises such as Topigs Norsvin; both use these boards to properly analyze scientific breakthroughs and their possible repercussions.
Several codes of conduct for responsible breeding, such as the industry-driven Code-EFABAR, need frequent modifications to incorporate new technology. Engaging diverse stakeholders in ethical discussions may provide a solid framework for these improvements. Animal ethics goes beyond well-being and requires thoroughly examining various issues to inform breeding choices and moral norms. Breeding groups and enterprises should explore the more significant ethical implications of GM and genome editing in cattle, ensuring the public that these concerns are handled appropriately.
The Bottom Line
As we’ve explored, genetic modification and genome editing in dairy cattle breeding are complex yet revolutionary. They offer the potential for disease-resistant, productive, and eco-friendly livestock to meet rising global dairy demand. However, ethical considerations must prioritize animal welfare, sustainability, and biodiversity. Science and ethics should inform each other, and dairy farmers or breeders must adopt best practices and make informed, ethical decisions. Genome editing can significantly contribute to a balanced and sustainable dairy industry with transparency, responsible use, and thoughtful discussion.












