Successful dairy cattle breeding is about using the facts available including the degree of trust that can be placed in the numbers. The facts used by breeders can vary all the way from in-herd observations, to show results, to including actual performance and genetic evaluation indexes. This article will deal with the genetic evaluation indexes that are based to a great extent on an animal’s DNA analysis. Often just referred to as ‘genomics.’ In this article, The Bullvine will cover details, from recently released studies and articles. We will look at how genomic evaluations are adding trustworthy information to the toolkit that breeders can use to advance their herds genetically.
1) Accuracy
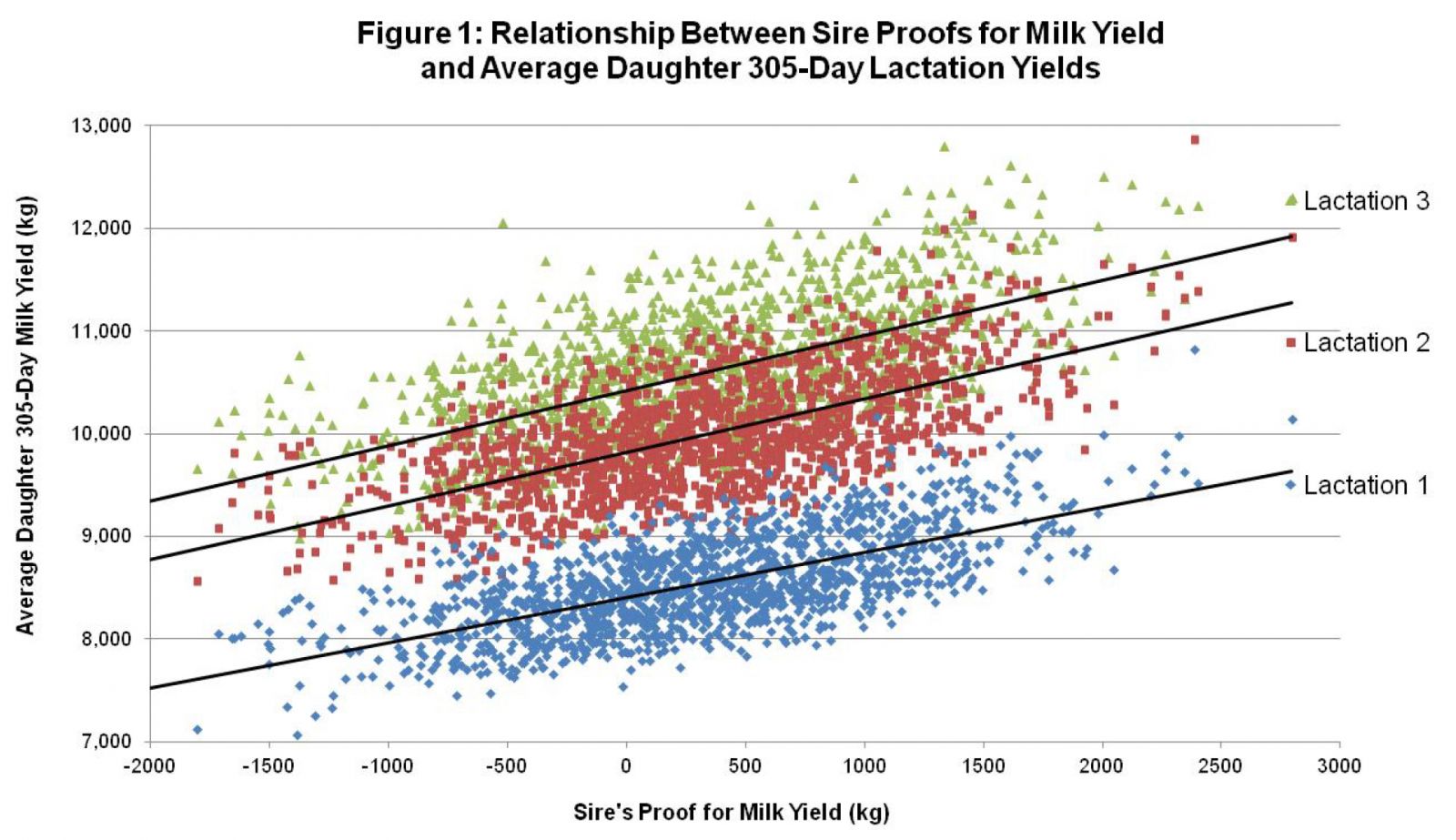
Before there were genomic indexes, there were parent average genetic indexes (PA’s) for heifers that did not have their performance (production and type) records of for bulls that did not have daughters with a performance recorded. The prediction accuracy for PA’s was low, standing at 20-33%. Breeders knew that there would be a wide variation from the PA numbers, once performance data was added in.
In 2008, based on the study of the DNA profiles of daughters proven sires, genomic (genetic) indexes were published by genetic evaluation centers that used both pedigree performance information and an animal’s DNA profile. Immediately the accuracy of the genomic indexes doubled (60-65%) those for PA’s. Of course, this was lower than the accuracies for extensively daughter proven sires, but a significant step forward.
Alta Genetics has recently published an excellent article on the accuracy of genomic index predictions – “How genomic proofs hold up.” The study compares genomic indexes at the time of release as young sires and what their indexes are in April 2017.
The study reports:
- Young sires released in 2010 2014 decreased by 171 vs. 52 in TPI and by 151 vs. 74 in NM$.
- For the 1078 US A.I. Holstein bulls released in 2013, their April 2017 indexes decreased on average by 99 TPI and 103 NM$. The degrees of change for TPI were: 4% of bull lost more than 300 TPI points; 9% remain, in 2017, within 20 TPI points of their 2013 indexes; and 19% increased in TPI from their 2013 to 2017 indexes. For NM$: 2% on the bulls changes by more than 300 in NM$; 9% were within 20 NM$ in 2017 of their 2013 indexes, and 9% increased in their NM$ index.
Definitely, there was an increase in accuracy of prediction of genomic indexes from 2010 to 2017.
Take Home Message: With each passing year, breeders can place more and more trust in the accuracy of genomics indexes. As more animals have their DNA profile established and as more SNIP research is conducted breeders can expect to see further increases in accuracy of genomic indexes. Also, there will, in the future, be the publication for additional genomic indexes for specific fats and proteins, for lifetime performance and for health and fertility traits.
2) Improvement Rates
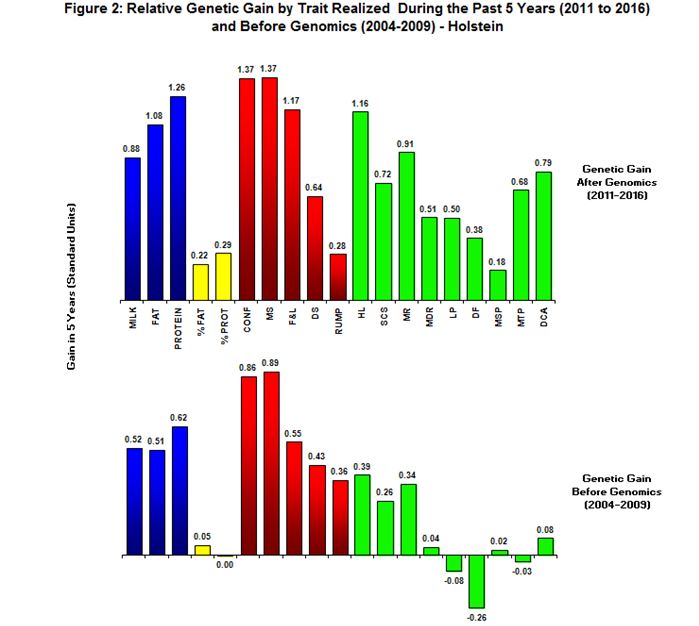
CDN has recently reported on a study “Analysis of Genetic Gains Realized Since Genomics.” This study compares two five-year time periods: (a) animals born (2004-2009) immediately prior to the existence of genomic evaluations; and (b) animals born (2011-2016) after genomic evaluations were available to breeders.
The rates vary by trait with the range in compared indexes being from a small improvement rate to over 500%. Note that in Holsteins the rate of genetic gain in protein %, lactation persistency (LP), daughter fertility (DF) and milking temperament (MTP) went from negative to positive. In Jerseys LP, MSP and daughter calving ability (DCA) went from negative to positive, yet metabolic disease resistance (MDR) went slightly negative. Similar rates of improved genetic gains were achieved by both Ayrshire and Brown Swiss breeds.
Take Home Message: Congratulations to the breeders for trusting and using the genomic index information to make faster rates of genetic improvement. A word of thanks goes out to the genetic evaluation centers all over the world for doing the research on and implementation of genomic indexes. The very significant increased rates of genetic gain may not be duplicated in the future for all traits as breeders are now selecting for many new economically important traits not previously evaluated and published.
3) Terminology
It is a known fact that the term ‘genomics’ has not always been interpreted correctly by everyone.
Over forty years ago, when genetic indexes were first published, frequently breeders thought of them as only being for production traits when they were available for both production and type traits. Today many people refer to genomic indexes as only being for production traits when they are available for production, type, fertility, health, other functional traits and total merit indexes (TPI, NM$, …).
Take Home Message: Interpret genomic indexes to be genetic indexes that include both pedigree and DNA profile information. Breeders can find genomically evaluated sires for all traits at all A.I. studs. Breeders can use one or all the genomic indexes as part of their herd’s breeding plan.
4) Inbreeding
Alta Genetics recently published an article, “Inbreeding: Manage it to Maximize Profit,” on sire options to limit the effects of inbreeding.
The article covers:
- When selection is practiced in a population, it results in a concentration of good genes and thus inbreeding. So, inbreeding is a natural outcome of selecting the best and eliminating the rest.
- Every 1% increase in inbreeding results in $22 – $24 less profit over a cow’s lifetime.
- There is not a magic level of inbreeding to be avoided. The current average level of inbreeding in North American Holsteins is 7-8%.
- A Midwest US study shows that superior inbred high genetic merit cows are more profitable than inferior genetic merit non-inbred cows.
The average inbreeding level of the top 25 NM$ (April ’17) daughter proven Holstein sires is 7.9% for genomic future inbreeding index (GFI). For the top 25 NM$ genomically evaluated sires the average GFI is 8.2%. Having genomic bulls with a higher level of inbreeding than proven sires is as expected when selection pressure is high, when generations are turned rapidly and when there is extensive focus placed on a single total merit indexes (NM$ or TPI or Pro$ or LPI or …).
Take Home Message: A.I. sire mating programs are designed to take into consideration the level of inbreeding of future progeny when a sire x dam is recommended. If a Holstein sire has a GFI of 9% or higher a breeder should require that that bull should have positive proof values for all of DPR, HCR, CCR, LIV, PL, SCC, immunity and calf wellness. Breeders should use and trust that inbreeding is being handled by sire mating programs.
5) Functional Traits
At the same time, as genomic evaluations became available, breeders started paying attention to a host of functional traits. These traits have economic significance and include milk quality, fertility, heifer, and cow health (immunity, wellness, disease resistance, livability, …), birthing, productive life and mobility. In the future, these functional traits will be expanded as on-farm data, and DNA profiling on animals are recorded and farm data is sent to data analysis centers. Noteworthy is the fact that animal wellness and welfare will be front and center for consumers of dairy products.
Take Home Message: Breeders can trust in the published genetic evaluations for functional traits as animal DNA profiles play a significant role in increasing the prediction accuracies from 15-25% to 60-70%. Functional trait improvement will require that breeders pay attention to both genetic and farm management.
6) Feed Efficiency
Feed accounts for 50-60% input costs for heifers and cows on dairy farms. Any gains that can be made by selecting genetically superior animals for their ability to convert feedstuffs to milk and meat have the potential for breeders to make more profit.
Research and data analysis are underway or nearing completion in many countries including US, Netherlands, and Canada on using DNA data combined with nutrition trial data to produce genomic indexes for feed efficiency. Other trials are underway to electronically capture on-farm data on feed intake, dry matter intake (DMI). It is a well-established fact that level of production is highly correlated to DMI.
CDAB has just published that “AGIL/USDA has demonstrated the feasibility of publishing national genomic evaluations for residual feed intake (RFI) based on the data generated by the 5-year national feed intake project funded by USDA National Institute of Food and Agriculture (NIFA), involving several research groups”. “The next step for CACB is to develop a proposal on how to collect data for use in genetic analysis for feed efficiency.”
Take Home Message: There will be genomic indexes for feed efficiency likely with 2-3 years. Once again breeders will have a tool they can trust into breed animals that return more profit.
7) Breeder Acceptance
A.I.’s are reporting that 60 to 90% of their semen sales are from genomically evaluated bulls. That fact on its own says that breeders purchasing larger volumes of semen are putting their trust in genomic evaluations. However, breeders wanting daughter proven sire proofs need to be given that option provided they are prepared to pay extra for their semen.
Take home message: Breeders check books tell the whole story – Genomic Evaluations are trusted.
The Bullvine Bottom Line
In less than a decade the use of DNA data in genetic evaluations has gone from unknown and not understood to a trusted source of very useful information. Having genomic indexes has given breeders the opportunity to advance their breeding programs, their herds, and their on-farm profits. Trust in information is important to dairy cattle breeders and they have and will continue, in the future, to place their trust in genomic indexes.
Get original “Bullvine” content sent straight to your email inbox for free.
[related-posts-thumbnails]










.jpg)










Leave a Reply
You must be logged in to post a comment.